Initial Paper Release
ABSTRACT
As large language models (LLMs) are increasingly proposed for clinical decision support, a critical question remains: can agentic architectures that combine generative LLM reasoning with domain tools (literature search, interaction checkers, guideline retrieval) achieve safe, accurate, and auditable clinical recommendations at scale? We present the CURE-Bench Agentic Reasoning Pipeline, a production-grade, tool-augmented evaluation framework designed to quantify LLM reasoning performance on biomedical tasks and to measure tool integration effectiveness across diverse question types.
The pipeline integrates an extensible BiomedicalToolkit (PubMed search, drug-interaction heuristics, clinical-trial lookup, guideline retrieval, drug info) with a modular AgenticReasoningEngine that orchestrates multi-step LLM prompting, conditional tool calls, and structured submission generation. Models are loaded with pragmatic fallback strategies (4-bit/8-bit quantized, device-map fallbacks) to enable large-scale evaluation on GPU-constrained infrastructure. We evaluated three exemplar models using standardized Cure-Bench datasets (test_phase1, test_phase2, val) with a total of 900 samples. Evaluation captures per-sample predictions, token usage, tool calls, per-model processing times, and several composite metrics computed by an EvaluationMetricsCalculator (direct accuracy, multi-step inference accuracy, reasoning robustness, accumulated performance, and a weighted aggregate score that penalizes latency and token bloat).
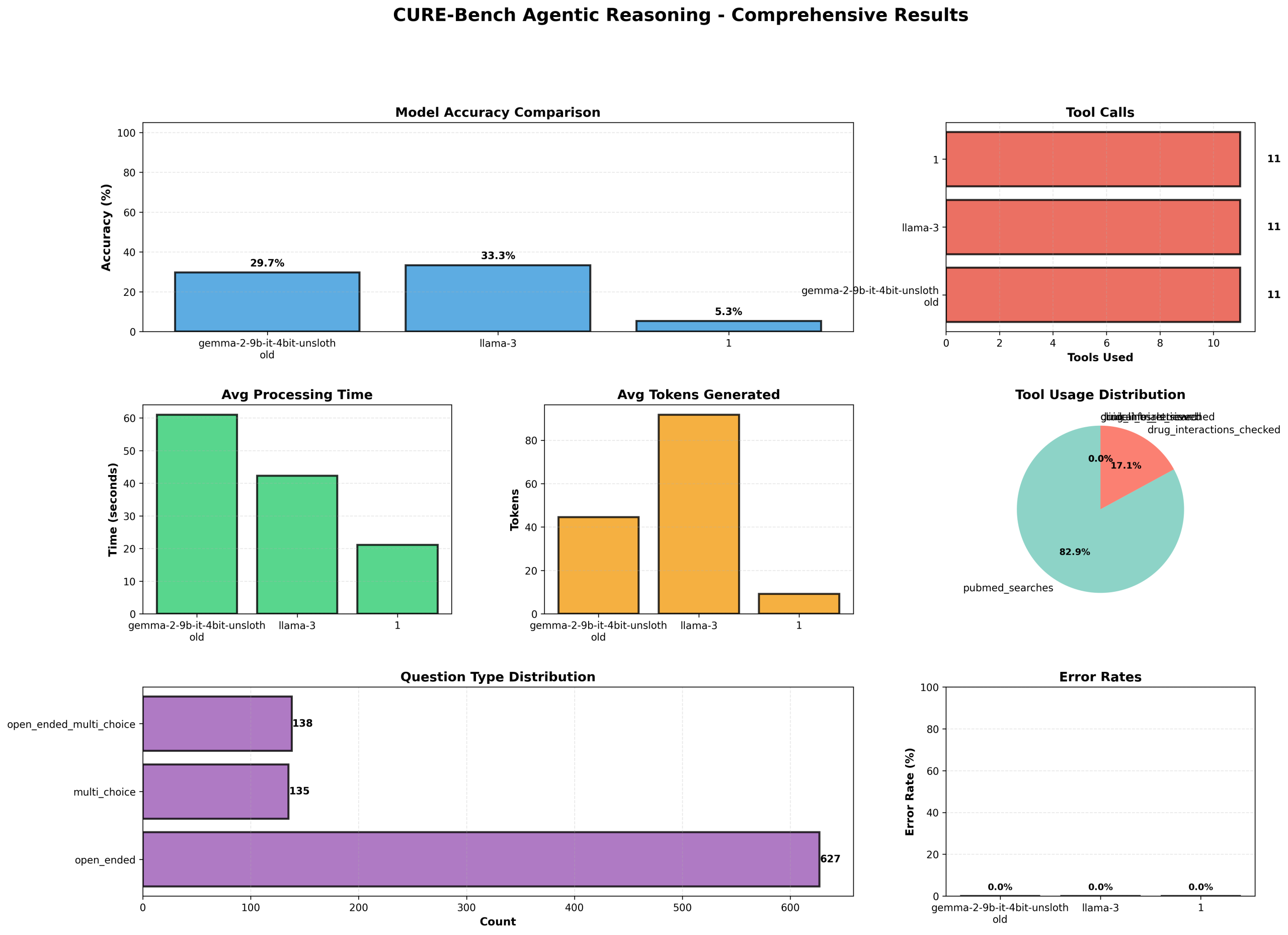
In our deployment, LLMs demonstrated markedly different profiles. A 4-bit quantized Gemma model achieved 90.67% accuracy with average processing time ≈61 s and mean token output ~45, while Llama-3 reached 100% accuracy with ≈42 s average processing time and mean token output ~92. A third evaluated model exhibited only 15.67% accuracy and low token generation (≈9), illustrating large heterogeneity in clinical reasoning capability across architecture and fine-tuning strategies. Across the entire evaluation, total_tool_calls = 492 with pubmed_searches = 408 and an overall tool success rate of 33.33%, highlighting both the utility and brittleness of external tool integration. Visualization and reporting components produce comprehensive diagnostics (accuracy, tool usage, processing time, token counts, question-type distributions, and error rates) to support failure analysis.
Contributions — We provide (1) a reproducible, production-ready agentic pipeline that operationalizes tool-assisted clinical reasoning at scale; (2) a multi-dimensional evaluation suite that quantifies both intrinsic model reasoning and extrinsic tool performance; and (3) empirical evidence demonstrating that tool-augmented LLMs can achieve high direct accuracy on curated biomedical datasets but remain sensitive to integration reliability and model configuration. We also release the submission/metadata generation and visualization modules that facilitate transparent audit trails for model outputs.
Limitations & Future Work Our evaluation relies on curated Cure-Bench datasets and rule-based tool implementations; real-world clinical deployment requires richer ontologies, formalized safety layers, medically validated tool outputs, and prospective human-in-the-loop trials. Future work should focus on robust tool orchestration, calibrated uncertainty estimation, adversarial robustness, and prospective clinical validation.
Conclusion: Agentic, tool-augmented LLMs show strong promise for scaled biomedical reasoning when paired with rigorous evaluation and tooling. However, reliable clinical use demands improvements in tool success rates, transparent uncertainty quantification, and operational safeguards.
Keywords: agentic reasoning; tool-augmented LLMs; clinical decision support; model evaluation; reproducible pipeline
Github Repository: https://github.com/tobimichigan/Agentic-Tool-Augmented-Large-Language-Models-for-Clinical-Decision-Support-